Family: Amnoonviridae
Eugene V. Koonin, Mart Krupovic, Win Surachetpong (วิน สุรเชษฐพงษ์), Yuri I. Wolf, and Jens H. Kuhn
The citation for this ICTV Report chapter is the summary to be published as Koonin et al., (2023):
ICTV Virus Taxonomy Profile: Amnoonviridae, Journal of General Virology 2023, (in press)
Corresponding author: Jens H. Kuhn (kuhnjens@mail.nih.gov)
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: September 2023
Summary
Amnoonviridae is a family of negative-sense RNA viruses with genomes totaling about 10.3 kb (Table 1.Amnoonviridae). These viruses have been found in actinopterygiid fish (in particular tilapia) and squamate reptiles (Gulf tree gehyras) sampled in Africa, Asia, and North America. The family includes a single genus with one species for one virus. The amnoonvirid genome consists of 10 segments, each with at least one open reading frame (ORF). The RNA1–3 ORFs encode the three subunits of the viral polymerase. The RNA4 ORF encodes a nucleoprotein.
Table 1.Amnoonviridae Characteristics of members of the family Amnoonviridae
| Characteristic | Description |
| Example | tilapia lake virus (KU751814–KU751823), species Tilapinevirus tilapiae, genus Tilapinevirus |
| Virion | Enveloped spherical particles of 55–100 nm |
| Genome | About 10.3 kb of deca-segmented negative-sense RNA |
| Replication | Unknown |
| Translation | Unknown |
| Host range | Actinopterygiid fish and squamate reptiles |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Insthoviricetes, order Articulavirales; the family includes one genus and one species. |
Virion
Morphology
Amnoonvirids produce spherical, slightly pleomorphic and likely enveloped particles that are 55–100 nm in diameter (Eyngor et al., 2014, Del-Pozo et al., 2017).
Nucleic acid
Amnoonvirids have 10 segments of linear negative-sense RNA with a total length of about 10.3 kb (tilapia lake virus - RNA1: 1,641 nt; RNA2: 1,471 nt; RNA3: 1,371 nt; RNA4: 1,250 nt; RNA5: 1,099 nt; RNA6: 1,044 nt; RNA7: 777 nt; RNA8: 657 nt; RNA9: 548 nt; and RNA10: 465 nt (Bacharach et al., 2016).
Genome organization and replication
Viruses of the family Amnoonviridae have deca-segmented genomes, with each segment possessing one primary ORF (Figure 1.Amnoonviridae). The RNA1 ORF encodes a protein with an RNA-directed RNA polymerase (RdRP) domain homologous to influenza C virus (Orthomyxoviridae: Gammainfluenzavirus) PB1 (Bacharach et al., 2016, Acharya et al., 2019, Ortiz-Baez et al., 2020). The RNA2 and RNA3 ORFs encode the PB2 and PA polymerase subunit homologs of orthomyxovirids (Arragain et al., 2023). The RNA4 ORF encodes a nucleoprotein (Abu Rass et al., 2022b). The identities and functions of proteins encoded by the remaining segments are unknown. The replication cycle of amnoonvirids remains to be determined. All 10 segments have conserved, complementary sequences at their 5′- and 3′-ends, similar to segment termini found in orthomyxovirid genome sequences (Bacharach et al., 2016). In contrast to orthomyxovirids, amnoonvirids do not hemagglutinate red blood cells, suggesting that they enter host cells differently (Chengula et al., 2019). However, entry occurs via dynamin-mediated endocytosis in a cholesterol-dependent, cytoskeleton-dependent manner that is independent of clathrin and pH (Abu Rass et al., 2022a).
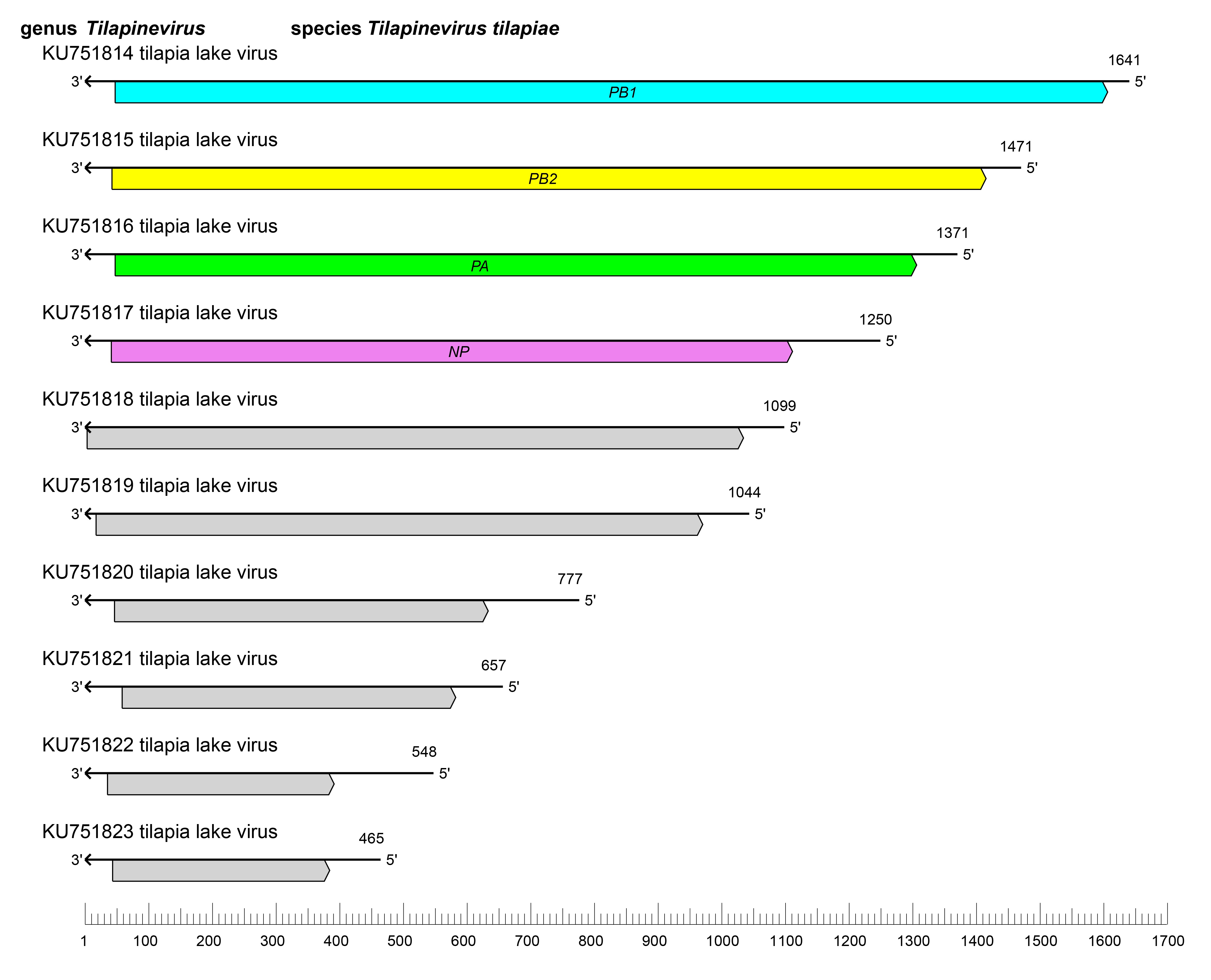 |
| Figure 1.Amnoonviridae. Genome length, organization, and position of open reading frames (ORFs) of tilapia lake virus, the only classified virus in the family Amnoonviridae, ORFs are indicated as boxes, colored according to the predicted protein function (NP, nucleoprotein gene; PA; polymerase subunit 3 gene; PB1, polymerase subunit 1 gene encoding an RdRP domain; PB2, polymerase subunit 2 gene). Genes of unknown function are colored gray. |
Biology
The only classified amnoonvirid, tilapia lake virus (TiLV), has been detected in sick and deceased wild and farmed Nile tilapia (cichlid Oreochromis niloticus (Linnaeus, 1758)), Nile tilapia x blue tilapia (Oreochromis aureus (Steindachner, 1864)) hybrids, and Mozambique tilapia (Oreochromis mossambicus (W. K. H. Peters, 1852)) in Bangladesh (Debnath et al., 2020), China/Taiwan (World Organisation for Animal Health 2023), Colombia (Contreras et al., 2021), Ecuador (Bacharach et al., 2016), Egypt (Nicholson et al., 2017), India (Swaminathan et al., 2021), Israel (Bacharach et al., 2016), Kenya (Mugimba et al., 2018), Malaysia (World Organisation for Animal Health 2023), Mexico (World Organisation for Animal Health 2023), Peru (World Organisation for Animal Health 2023), Philippines (World Organisation for Animal Health 2023), Tanzania (Mugimba et al., 2018), Thailand (Surachetpong et al., 2017), Uganda (Mugimba et al., 2018), United States (Ahasan et al., 2020), and Viet Nam (Tran et al., 2022). TiLV has also been found in apparently healthy giant gourami (osphronemid Osphronemus goramy Lacépède, 1801) sampled in Thailand (Chiamkunakorn et al., 2019).
Unclassified amnoonvirids have been found in diverse actinopterygiid fish (Turnbull et al., 2020, Li 2023), with the exception of Lauta virus, which was discovered in Gulf tree gehyras (gekkonid Gehyra lauta Oliver, Prasetya, Tedeschi, Fenker, Ellis, Doughty, & Moritz, 2020) sampled in Australia (Ortiz-Baez et al., 2020).
TiLV causes a systemic disease involving most organs, with virus present also in muscles, mucus, and feces (Eyngor et al., 2014, Bacharach et al., 2016, Tattiyapong et al., 2018, Mugimba et al., 2019, Yamkasem et al., 2021b). Infection results in lethargy, ocular disease (such as, cataracts, endophthalmitis or exophthalmos), skin erosions and hemorrhages, and severe anemia, leading to death in >80% of cases (Eyngor et al., 2014, Tattiyapong et al., 2017, Turner et al., 2023). TiLV is also virulent in experimentally exposed larval and adult zebrafish (cyprinid Danio rerio (F. Hamilton, 1822)) (Rakus et al., 2020, Widziolek et al., 2021), Malawi cichlids (Aulonacara spp.) (Yamkasem et al., 2021a), and Mozambique tilapia (Oreochromis mossambicus (W. K. H. Peters, 1852)) (Waiyamitra et al., 2021), and replicates subclinically in firemouth cichlids (Thorichthys meeki Brind, 1918) and freshwater angelfish (cichlid Pterophyllum scalare (Schultze, 1823)) (Paria et al., 2023). Experimentally, TiLV can infect and cause mass mortality in red tilapia and giant gourami, but does not infect Asian sea bass (latid Lates calcarifer (Bloch, 1790)), climbing perch (anabantid Anabas testudineus (Bloch, 1792)), common carp (cyprinid Cyprinus carpio Linnaeus, 1758), goldfish (cyprinid Carassius auratus (Linnaeus, 1758)), iridescent sharks (pangasiid Pangasianodon hypophthalmus (Sauvage, 1878)), parrotfish (cichlid Amphilophus citrinellus (Günther, 1864) x Vieja melanurus (Günther, 1862)), silver barb (cyprinid Barbodes gonionotus (Bleeker, 1850)), snakeskin gouramis (osphronemid Trichogaster pectoralis Regan, 1910), striped snake-head fish (channid Channa striata (Bloch, 1793)), three spot gourami (osphronemid Trichopodus trichopterus (Pallas, 1770)), or walking catfish (clariid Clarias macrocephalus Günther, 1864) (Jaemwimol et al., 2018, Paria et al., 2023).
Derivation of names
Amnoonviridae: from Hebrew אַמְנוּן [amnun/amnoon] for tilapia
tilapiae: from tilapia
Tilapinevirus: from the adjective tilapine, referring to fish of tribe Tilap(i)ini
Genus demarcation criteria
Not applicable (the family includes only a single genus).
Species demarcation criteria
Not applicable (the only genus includes only a single species).
Relationships within the family
Phylogenetic relationships of TiLV to members of the phylum Negarnaviricota are shown in Figure 2.Amnoonviridae.
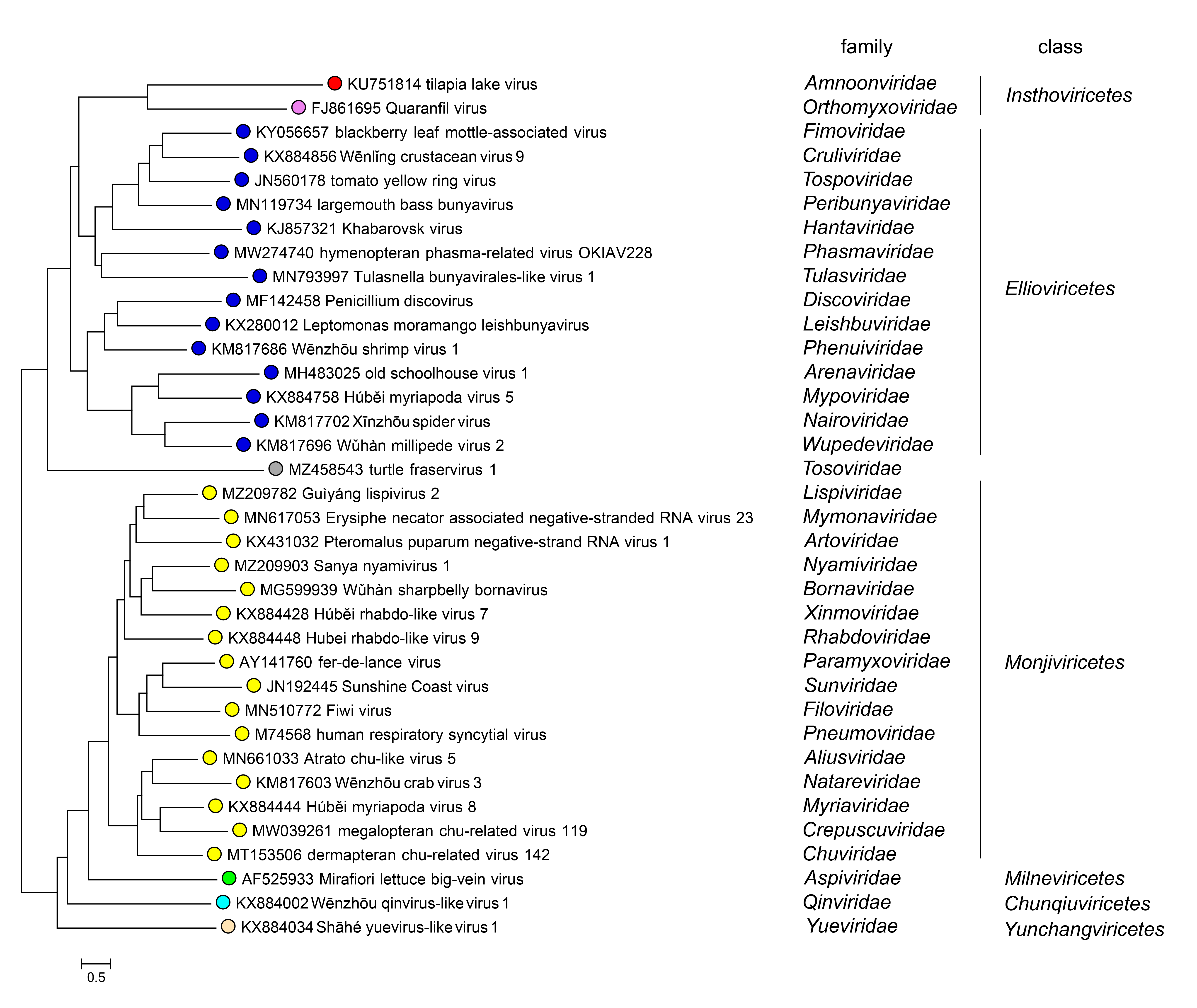 |
| Figure 2.Amnoonviridae. Phylogenetic relationships of TiLV to members of the phylum Negarnaviricota. The list of complete exemplar genomes of Negarnaviricota was obtained from the ICTV Virus Metadata Resource (VMR) datasheet v.38.1 (https://ictv.global/vmr). RdRP core domains were identified by running a PSI-BLAST search against either the protein coding gene complement or six-frame translation of the virus RNA sequences using the 143 negarnaviricot profiles (Neri et al., 2022) and a tosovirid profile (Waltzek et al., 2022). RdRP core fragments were clustered using MMSEQS2 with the sequence similarity threshold of 0.3; sequences in each cluster were aligned using MUSCLE5. A consensus sequence was derived from each alignment; consensus sequences were aligned using MUSCLE5, then each consensus amino acid was expanded into an alignment column (Neri et al., 2022). An approximate ML phylogenetic tree was reconstructed from this alignment of 1122 negarnaviricot RdRP core sequences using FastTree (WAG evolutionary model, gamma-distributed site rates). A representative sequence (one, closest to the family root) was selected for each of the 36 negarnaviricot families and the full tree was trimmed to these 36 representative leaves. |
Relationships with other taxa
Viruses in the family Amnoonviridae are most closely related to articulaviral orthomyxovirids (Bacharach et al., 2016).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| Lauta virus | MT386081* | LTAV | (Ortiz-Baez et al., 2020) |
Virus names and virus abbreviations are not official ICTV designations.
* Incomplete genome

