Family: Xinmoviridae
Stephen R. Sharpe and Sofia Paraskevopoulou (Σοφία Παρασκευοπούλου)
The citation for this ICTV Report chapter is the summary to be published as Sharpe and Paraskevopoulou (2023):
ICTV Virus Taxonomy Profile: Xinmoviridae 2023, Journal of General Virology, 104: 001906
Corresponding authors: Stephen Sharpe Stephen.Sharpe@csiro.au and Sofia Paraskevopoulou paraskevopoulous@rki.de
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: September 2023
Summary
Xinmoviridae is a family of viruses with negative-sense RNA genomes of 9–14 kb (Table 1.Xinmoviridae). The family includes 12 genera and 14 species. Xinmovirids typically infect beneficial and pest insects in Africa, Asia, Europe, and Oceania, but their host range and geographic distribution has not yet been investigated systematically and hence may be broader.
Table 1. Xinmoviridae. Characteristics of members of the family Xinmoviridae
| Characteristic | Description |
| Example | Drosophila unispina virus 1 (KR822819), species Drunivirus chambonense, genus Drunivirus |
| Virion | Not known |
| Genome | 9–14 kb of negative-sense RNA |
| Replication | Not known |
| Translation | Not known |
| Host range | Arthropoda |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Monjiviricetes, order Mononegavirales: 12 genera and 14 species |
Virion
Morphology
Xinmovirids are thus far only known from metagenomics studies. Virions have not yet been visualised and structural proteins have not been studied.
Genome organization and replication
Xinmovirids have negative-sense RNA genomes of 9–14 kb with three to six ORFs (Figure 1.Xinmoviridae) (Parry and Asgari 2018). These ORFs encode at least three structural proteins that have been identified via comparison with proteins encoded by other viruses in the order Mononegavirales: a glycoprotein, a nucleoprotein, and an RNA directed RNA polymerase (RdRP) (Scarpassa et al., 2019).
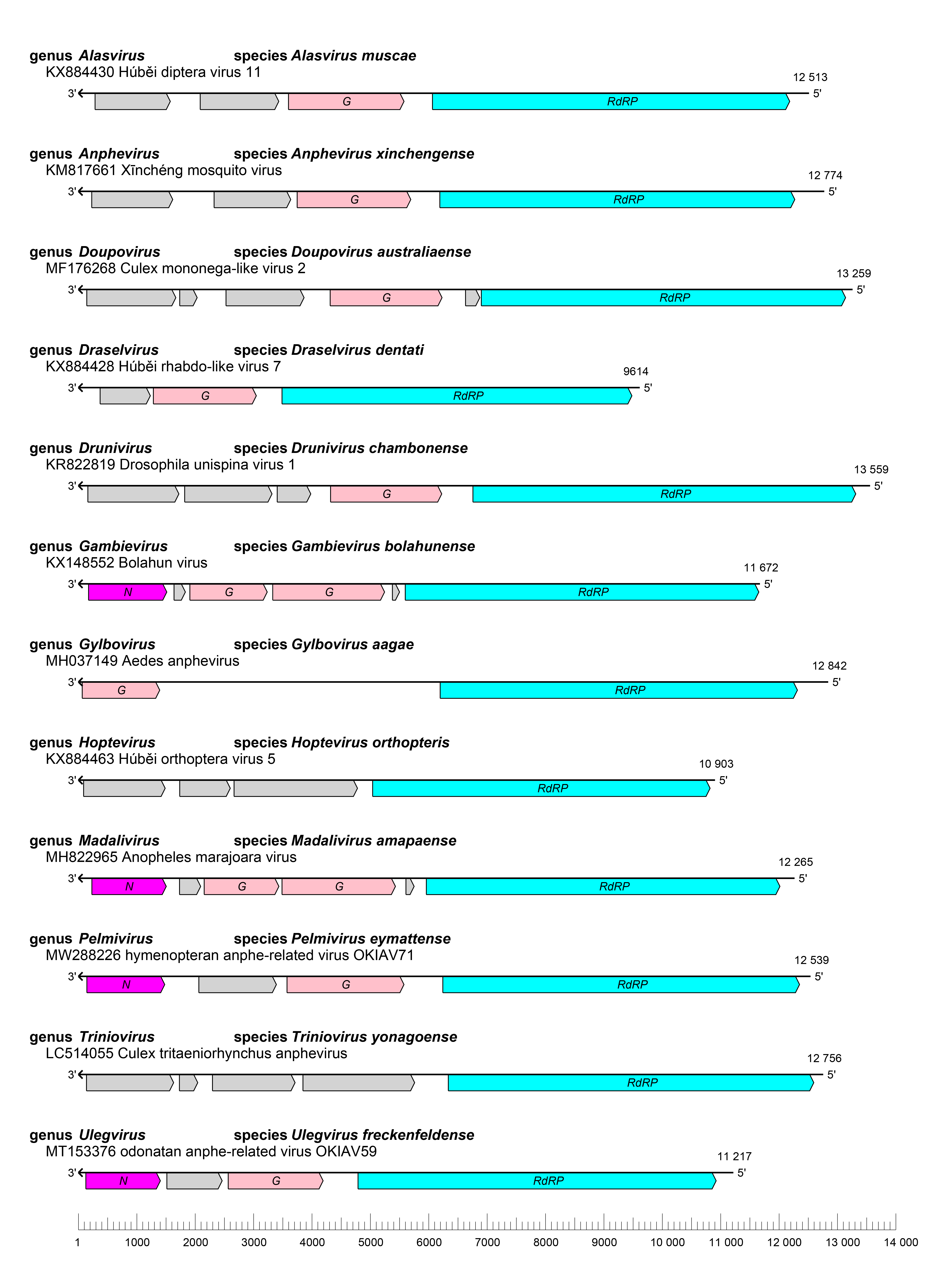 |
| Figure 1.Xinmoviridae. Genome organisation of representative viruses of each genus in the family Xinmoviridae. Boxes show the position of ORFs coloured according to their predicted protein function: N, nucleoprotein gene (magenta); G, glycoprotein gene (pink); RdRP, RNA-directed RNA polymerase gene (cyan). The GenBank accession number and length of each genome is included. |
Biology
Viruses in the family Xinmoviridae have been detected in insects from Africa (Tanzania), Asia (Japan), Europe (Croatia, Germany, Greece, Italy, Switzerland), and Oceania (Australia), and may infect insects of more than 12 orders, including Blattodea, Coleoptera, Diptera (Shi et al., 2017, Scarpassa et al., 2019), Hemiptera, Hymenoptera, and Lepidoptera, Mantodea, Neuroptera, Odonata, Orthoptera, Psocodea, Trichoptera, and Zygentoma (Käfer et al., 2019).
Derivation of names
Alasvirus: from the Latin alas meaning wings, referring to Húběi diptera virus 11, which was discovered by HTS in a pool of two-winged flies. The species epithet muscae derives from musca, the Latin word for fly.
Anphevirus: from the insect genus Anopheles, Xīnchéng anphevirus having been discovered by HTS in Anopheles sinensis mosquitoes. The species epithet xinchengense refers to Xīnchéng, China, the sample location.
Doupovirus: from Point Douro, Australia, where Culex mononega-like virus 2 was discovered by HTS in Culex australicus mosquitoes. The species epithet australiaense refers to Australia.
Draselvirus: from dragonflies and damselflies, odonates in which Húběi rhabdo-like virus 7 was discovered by HTS. The species epithet dentati derives from dentatum, the Latin word for toothed (odonate is derived from ὀδούς, the Greek word for tooth).
Drunivirus: from Drosophila unispina, the host in which Drosophila unispina virus 1 was discovered. The species epithet chambonense derives from Chambon, France, the sample location.
Gambievirus: from Gambie virus which was discovered by HTS in Anopheles gambiae mosquitoes sampled in Senegal (Gambie is the French name of Gambia). The species epithet bolahunense derives from Bolahun virus discovered by HTS in Anopheles gambiae mosquitoes sampled in Bolahun, Liberia. The species epithet senegalense derives from Senegal, the sampling location for Gambie virus.
Gylbovirus: from Aedes aegypti and Aedes albopictus, hosts or host cells associated with Aedes anphevirus infection. The species epithet aagae derives from the cell line Aag2.
Hoptevirus: from Orthoptera, the order to which insects in which Húběi orthoptera virus 5 was discovered by HTS are assigned. The species epithet orthopteris is derived from Orthoptera.
Madalivirus: from Anopheles marajoara and Anopheles darling, the species of mosquitoes in which viruses in the genus were first discovered. The species epithet amapaense derives from Amapá state, Brazil, the sampling location of Anopheles marajoara virus. The species epithet amazonaense derives from Amazonas state, Brazil, one of two mosquito sampling sites in Brazil where Anopheles darlingi virus was discovered by HTS.
Pelmivirus: from Heteropelma amictum, the species of parasitoid wasps in which hymenopteran anphe-related virus OKIAV71 was discovered by HTS. The species epithet eymattense refers to the sample site of Eymatt, Bern, Switzerland.
Triniovirus: from Culex tritaeniorhynchus, the species of mosquitoes in which Culex tritaeniorhynchus anphevirus was discovered by HTS. The species epithet yonagoense derives from Yonago, Japan, the site of sampling.
Ulegvirus: from Libellula and Cordulegaster, the genera of dragonflies in which viruses of this genus were discovered by HTS. The species epithet freckenfeldense derives from Freckenfeld, Rhineland-Palatinate, Germany, the sampling site.
Xinmoviridae: from Xīnchéng mosquito virus, the first virus that was assigned to the family.
Genus demarcation criteria
Members of different genera have RdRP amino acid identities of 60% or less.
Species demarcation criteria
Members of different species within a genus have RdRP amino acid identities of 66% or less.
Relationships within the family
Phylogenetic relationships of Xinmoviridae are shown in Figure 2.Xinmoviridae.
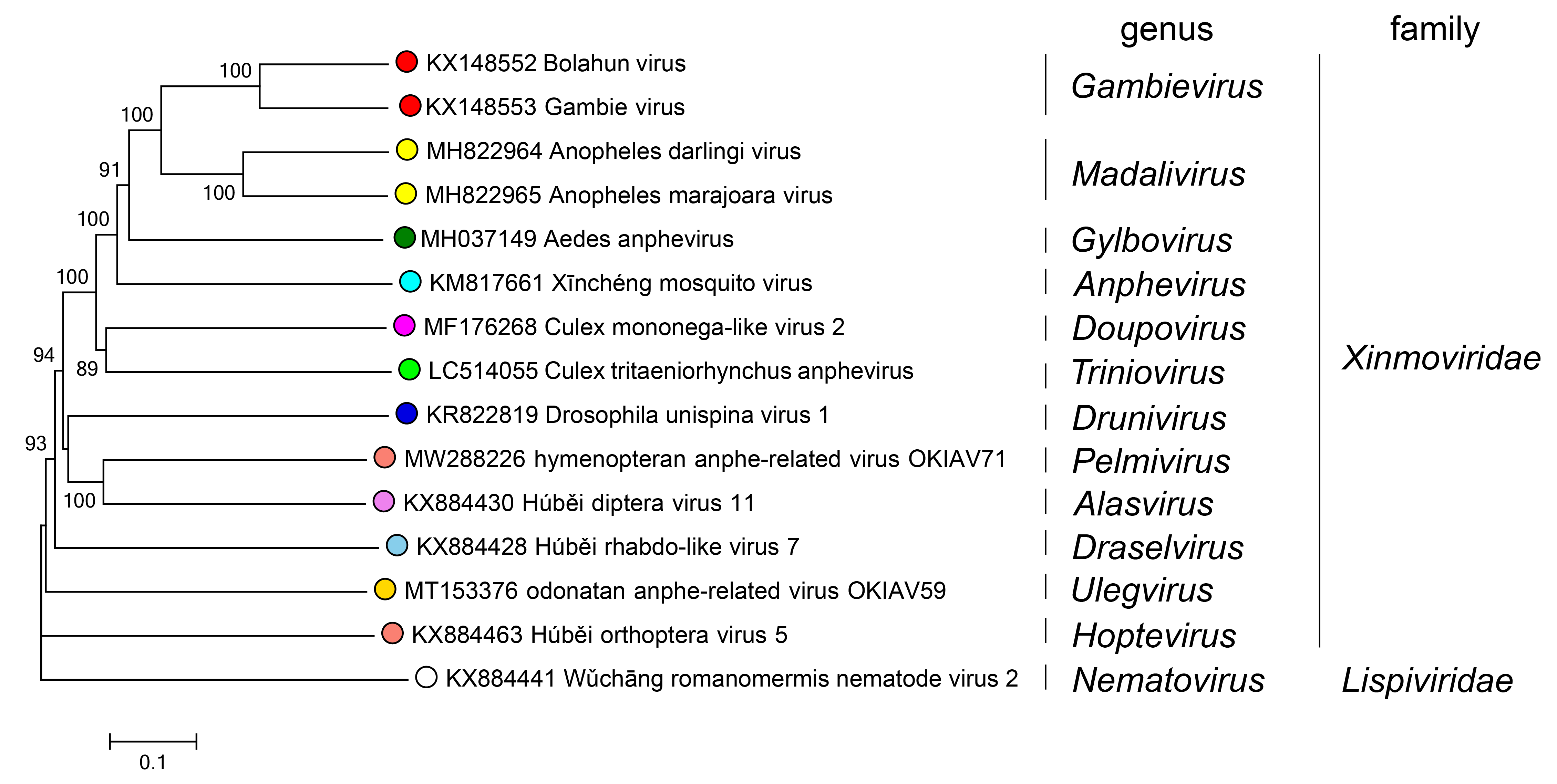 |
| Figure 2.Xinmoviridae. Phylogenetic tree of xinomovirid RdRP amino acid sequences. Sequences were aligned using MUSCLE v3.8 (Edgar 2004) and a neighbour-joining tree of amino acid distances was created using the Poisson correction within MEGA7 (Kumar et al., 2016). The virus Wǔchāng romanomermis nematode virus 2 (family Lispviridae) was used as an outgroup. Circles at tips are coloured according to genus. |
Relationships with other taxa
Viruses in the family Xinmoviridae are most closely related to members of the families Bornaviridae and Nyamiviridae, these families also being placed in the order Mononegavirales.
Related, unclassified viruses
Unclassified viruses that are likely to be members of the family Xinmoviridae have been detected in damselflies, leaf-beetles, mosquitoes (Shi et al., 2017, Manni and Zdobnov 2020, Stanojevic et al., 2020, Batson et al., 2021), shieldbugs, stinkbugs and tephritid fruit flies (Zhang et al., 2020, Sharpe et al., 2021). The table below includes only those viruses with complete or near-complete genome sequences.
| Virus name | Accession number | Virus abbreviation |
| Aedes aegypti anphevirus | MW452303 | |
| Aedes albopictus anphevirus | MW147277 | AealbAV |
| Bactrocera dorsalis xinmovirus 2 | HG994135 | BdXV2 |
| Bactrocera dorsalis borna-like virus | MN745081 | |
| bat faecal associated anphe-like virus 1 | ON872562 | |
| Culex mononega-like virus 2 strain mosWSX25901 | MF176377 | CMLV2 |
| Fushun monolepta lauta xinmovirus 1 | MZ210030 | |
| Fushun monolepta lauta xinmovirus 2 | MZ210031 | |
| Gordis virus | MW435019 | |
| Gudgenby calliphora mononega-like virus | MT129693 | |
| Guiyang xinmovirus 1 | MZ209642 | |
| Hangzhou cletus punctiger xinmovirus 1 | MZ209673 | |
| Hangzhou zicrona caerulea xinmovirus 1 | MZ209710 | |
| Sanya ischnura senegalensis xinmovirus 1 | MZ209914 | |
| Serbia mononega-like virus 1 | MT822181 |
Virus names and virus abbreviations are not official ICTV designations.

